Wednesday June 8, 2022

| Title | Value |
|---|---|
| Title | |
| Description | Leave empty |
| Directory |
| Parameter | Value |
|---|---|
| Movies data path | |
| Gain reference path | |
| Flip gain ref & defect file in Y? | Yes |
| Raw pixel size (A) | |
| Accelerating Voltage (kV) | |
| Spherical Aberration (mm) | |
| Total exposure dose (e/A^2) | |
| Skip Header Check | Yes |
| Parameter | Value |
|---|---|
| Number of GPUs to parallelize |
| Parameter | Value |
|---|---|
| Number of GPUs to parallelize |
Interactive job: accept all exposures below a CTF Fit Resolution (A) of 6.
| Parameter | Value |
|---|---|
| Minimum particle diameter (A) | |
| Maximum particle diameter (A) |
Interactive job: select picks with an NCC >= 0.2 and Power Score > 824 and < 2955
| Parameter | Value |
|---|---|
| Number of GPUs to parallelize (0 for CPU-only) |
| Parameter | Value |
|---|---|
| Number of GPUs to parallelize |
Interactive job
Use default parameters
| Parameter | Value |
|---|---|
| Symmetry |
Use the same parameters as the previous steps.
Select 'Apply to All' with all parameters set, enable the exposure group and start the session.
| Title | Value |
|---|---|
| Title | |
| Description | Leave empty |
| Directory |
| Parameter | Value |
|---|---|
| Movies data path | |
| Gain reference path | |
| Flip gain ref & defect file in Y? | Yes |
| Raw pixel size (A) | |
| Accelerating Voltage (kV) | |
| Spherical Aberration (mm) | |
| Total exposure dose (e/A^2) | |
| Skip Header Check | Yes |
| Parameter | Value |
|---|---|
| Number of GPUs to parallelize |
| Parameter | Value |
|---|---|
| Number of GPUs to parallelize |
| Parameter | Value |
|---|---|
| Minimum particle diameter (A) | |
| Maximum particle diameter (A) | |
| Use elliptical blob | Yes |
Interactive job: select picks with an NCC >= 0.2 and Power Score > 635 and < 1045
| Parameter | Value |
|---|---|
| Number of GPUs to parallelize (0 for CPU-only) | |
| Extraction box size (pix) | |
| Fourier crop to box size (pix) |
| Parameter | Value |
|---|---|
| Number of GPUs to parallelize |
Interactive job
Use default parameters
| Parameter | Value |
|---|---|
| Minimize over par-particle scale | Yes |
| Title | Value |
|---|---|
| Title | |
| Description | Leave empty |
| Directory |
To start processing, we will need to create a container for jobs to run. Select the 'New' button from the projects page and enter the following details:
| Title | Value |
|---|---|
| Title | |
| Description | Leave empty |
| Directory |
| Parameter | Value |
|---|---|
| Particle meta path |
| Parameter | Value |
|---|---|
| Circular mask diameter (A) |
Interactive job
| Parameter | Value |
|---|---|
| Number of Ab-Initio classes |
Use default parameters
| Parameter | Value |
|---|---|
| Target resolution (A) | |
| Number of O-EM epochs | |
| Number of final full iterations |
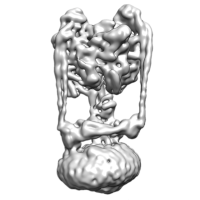
To start processing, we will need to create a container for jobs to run. Select the 'New' button from the projects page and enter the following details:
| Title | Value |
|---|---|
| Title | |
| Description | Leave empty |
| Directory |
| Parameter | Value |
|---|---|
| Particle meta path |
| Parameter | Value |
|---|---|
| Filter resolution (A) |
| Parameter | Value |
|---|---|
| Downsample to box size | |
| Crop to size (after downsample) |
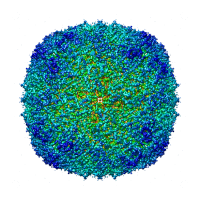
To start processing, we will need to create a container for jobs to run. Select the 'New' button from the projects page and enter the following details:
| Title | Value |
|---|---|
| Title | |
| Description | Leave empty |
| Directory |
| Parameter | Value |
|---|---|
| Particle meta path |
| Parameter | Value |
|---|---|
| Symmetry | |
| Initial lowpass resolution (A) | |
| Minimize over per-particle scale | Yes |
| Parameter | Value |
|---|---|
| Symmetry | |
| Initial lowpass resolution (A) | |
| Minimize over per-particle scale | Yes |
| Optimize per-particle defocus | Yes |
| Optimize per-group CTF params | Yes |